MINNEAPOLIS– At the 70th ASMS Conference, Bruker Corporation (Nasdaq: BRKR) announced key innovations for spatial multiomics of tissue and tumor microenvironments (TME). Following Bruker’s strategic partnership with AmberGen1, key enhancements are introduced for MALDI HiPLEX-IHC mass spectrometry imaging.
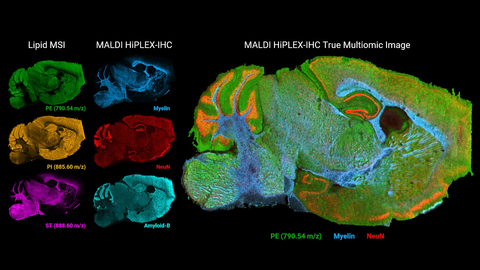
MALDI HiPLEX-IHC represents a breakthrough in multiomics imaging by combining targeted protein expression spatial profiling with unbiased small molecule MALDI Imaging to co-localize proteins and small molecules such as glycans, lipids, metabolites, or xenobiotics. Using AmberGen’s MiralysTM antibody-based photocleavable peptide mass tags, highly multiplexed IHC staining and photocleavage of peptide markers fit seamlessly into Bruker’s IntelliSlide®-based automated workflows for MALDI imaging.
Novel multiomic imaging enhances spatial high-plex protein imaging with the ability to elucidate metabolic processes in the same tissue section. In addition to mapping tens to over one hundred targeted proteins with high-plex peptide tags, MALDI HiPLEX-IHC can track signaling pathways such as glycosylation, observe lipid spatial profiles for tumor microenvironment segmentation, or simultaneously observe how drugs affect both protein and metabolic states.
Dr. Peggi Angel, Professor of Cell and Molecular Pharmacology and Experimental Therapeutics at the Medical University of South Carolina commented: “From the perspective of a lab heavily invested in cellular signaling processes in cancer biology, MALDI HiPLEX-IHC is a game changer allowing integration of mass spectrometry imaging with cell biology. We will be using this technology for multiomic N-glycan and collagen imaging studies to understand aggressive breast cancers. I expect that researchers investigating the tissue microenvironment will quickly adopt this unique spatial multiomics technology.”
Bruker also announced its microGRID module for smartbeam 3D MALDI sources for timsTOF fleX systems. The microGRID improves the MALDI stage to sub-micron precision to correct laser positioning on tissue surfaces down to 5 micrometers (µm), virtually eliminating any visual artifacts or artifacts in co-registration of MALDI images with optical microscopy. As correction is effective for entire pathology slides, microGRID leverages a large field-of-view for MALDI HiPLEX-IHC protein expression profiling.
Dr. Ron Heeren, Distinguished Professor and Limburg co-chair of the Maastricht Multimodal Molecular Imaging (M4I) Institute, added: “At M4I, we develop workflows and techniques to contextualize the role of individual cells in disease, and determine how interactions between cells affects cellular states locally and across long distances.”
Dr. Heeren continued: “Since we work closely with pathologists and cancer researchers who are used to microscopes, we cannot afford artifacts in our mass spectrometry images, and need large fields-of-view. Bruker’s microGRID achieves this effortlessly without limiting the area of the slide we can process for multiomic, multimodal images of diseased and homeostatic pathology.”
Bruker also demonstrated key updates for MALDI Imaging data analysis using SCiLSTM Lab 2023a, including microGRID support and improvements in handling CCS (collisional cross section)-enabled imaging datasets acquired with timsTOF technology. SCiLSTM Lab 2023a introduces the novel 4D Feature-Finder for visualizing CCS-resolved features in intuitive mass-mobility diagrams. In addition, images produced from multimodal datasets were demonstrated with MALDI HiPLEX-IHC protein expression spatial profiling. This allows SCiLS Lab to leverage its auto-segmentation and statistical profiling tools using images that overlay protein and small molecule localization on the same tissue section.

